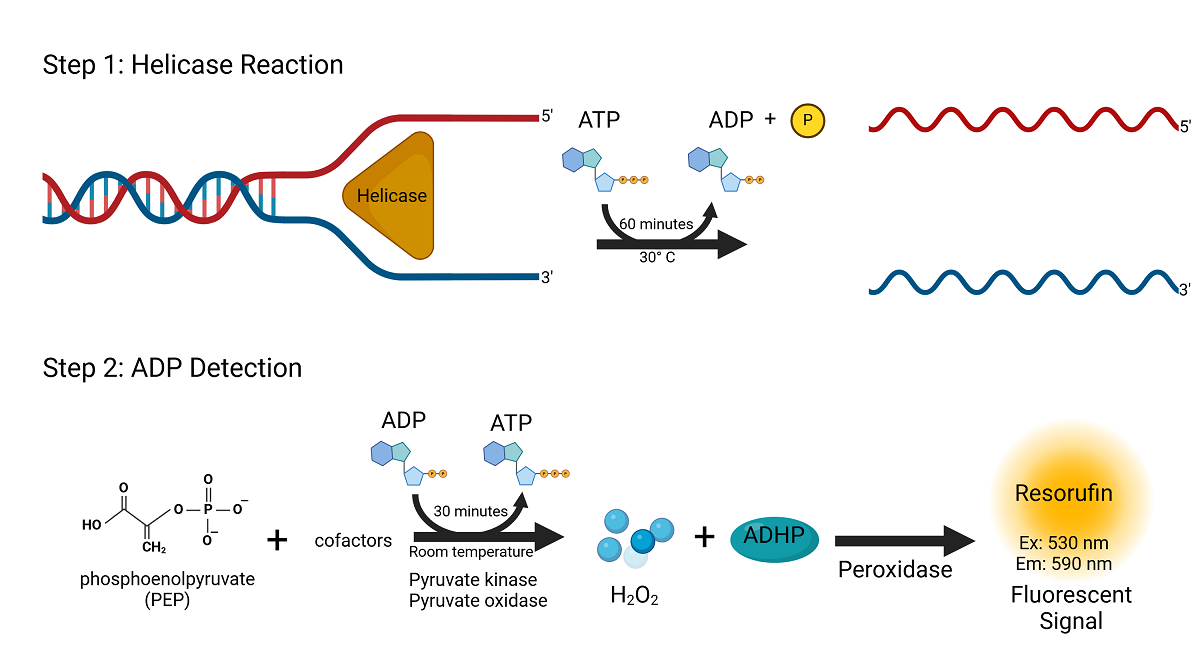
Figure 1. ATP consumption by helicases during DNA or RNA unwinding Assay Principle
This assay principle, initially developed for kinase measurements, is equally suited to detect ATP consumption by helicases during DNA or RNA unwinding. Our ADP Hunter Plus detection method has been successfully applied to validate the ATPase activity of recombinant helicases, ensuring robust performance in both drug discovery and mechanistic studies.
This homogeneous (no wash), gain-of-signal assay is non-radioactive and provides results in 90 minutes. The simple two-step procedure uses a coupled enzyme reaction system that generates hydrogen peroxide (H₂O₂) from ADP. Hydrogen peroxide, when combined with ADHP (fluorescent dye precursor) in the presence of peroxidase, generates a fluorescent active resorufin dye. A reaction stop solution is provided with the kit for added signal-and-background stabilization to allow for automation.
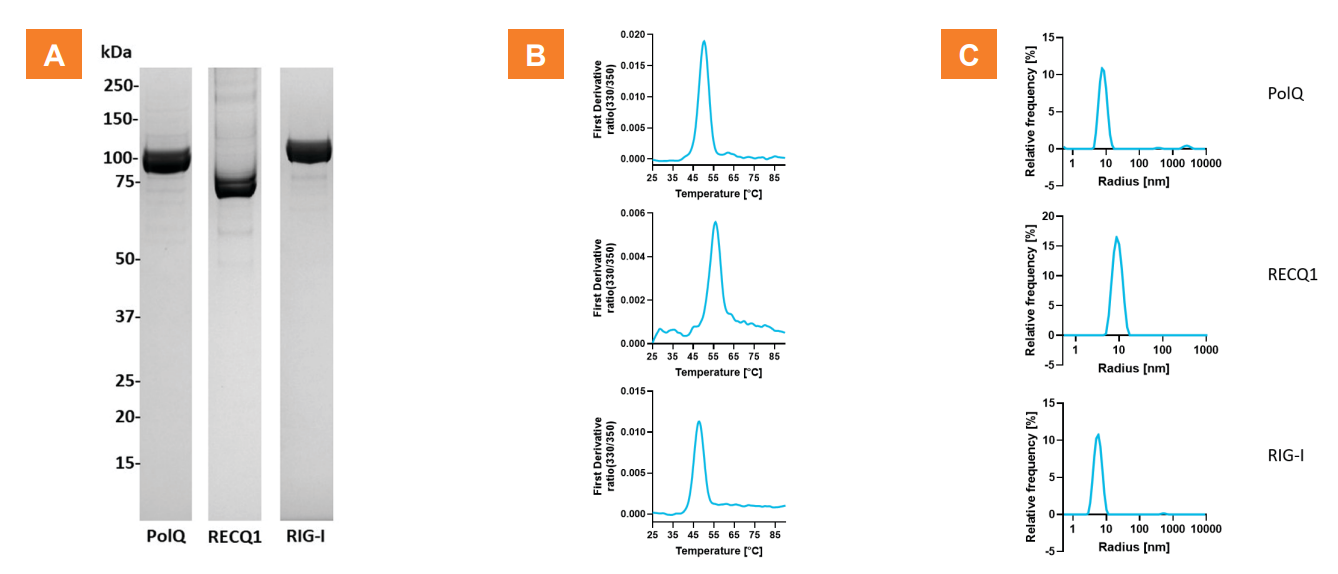
Figure 2. Helicase protein production and purity.
A. Analysis of the purified proteins by SDS-PAGE. The final samples are highly
pure as observed by SDS-PAGE with Coomassie Blue staining. Predicted molecular weights: PolQ (1-894): 104 kDa, RECQ1: 78
kDa, RIG-I: 111 kDa. B. Stability of purified PolQ, RECQ1 and RIG-I enzymes assessed by intrinsic fluorescence-based NanoDSF. The
proteins are folded with apparent melting temperatures of Tm PolQ = 55.9°C, Tm RECQ1 = 50.3°C, Tm RIG-I = 47.7°C. C. Dynamic
Light Scattering (DLS) analysis indicates that recombinant proteins are monodisperse with polydispersity indexes (PDI) of: 0.06±0.01,
0.07±0.02, and 0.14±0.01 for PolQ, RECQ1 and RIG-I respectively.
 Eurofins Discovery
Eurofins Discovery


 Eurofins Discovery
Eurofins Discovery